

On May 9, 2024, Professor Wang Xinghuan's team from Zhongnan Hospital of Wuhan University published a research paper titled "Tracking single cell evolution using clock like chromatin accessibility locality" in Nature Biotechnology (Top of District 1, Chinese Academy of Sciences, five-year IF=59.1).

At present, time series analysis of single-cell data usually requires specific experimental methods, such as using chemical markers, deep somatic mutation sequencing, lineage-specific reverse genetic markers, multi-time point sampling, single-cell methylation sequencing, etc. In order to facilitate the reconstruction of accurate developmental trajectories from any high-throughput single-cell sequencing dataset, it is necessary to identify intrinsic biomarkers within the data that can be used to report cell generation time. The research group and Professor Zhang Yi's team from Beijing Youle Fusheng have independently developed a traceability technology based on clock like site chromatin accessibility to infer single-cell replication age (EpiTrace), which has achieved cell age inference for single-cell chromatin accessibility (ATAC-seq) datasets and provided a time reference frame for this type of single-cell sequencing dataset.
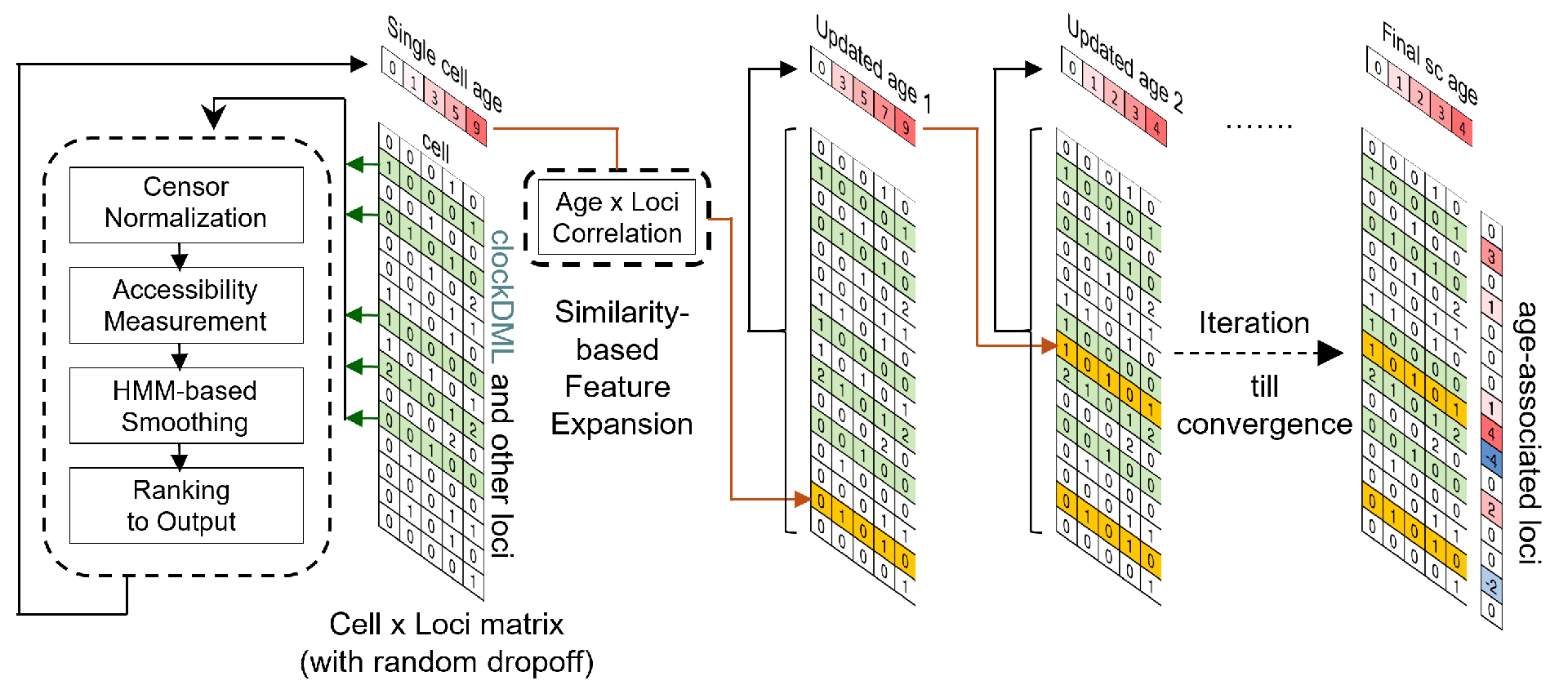
EpiTrace has good complementarity with RNA rate, mutation tracking, DNA methylation age inference, and multi-time point sampling. The research team used EpiTrace to track the key nodes in the development of bladder cancer, namely, TM4SF1 positive cancer cell subsets (TPCS), and found the key driving factors for the development of bladder cancer[2] (see Adv Sci | key nodes of epigenetic evolution of heterogeneity in bladder cancer).

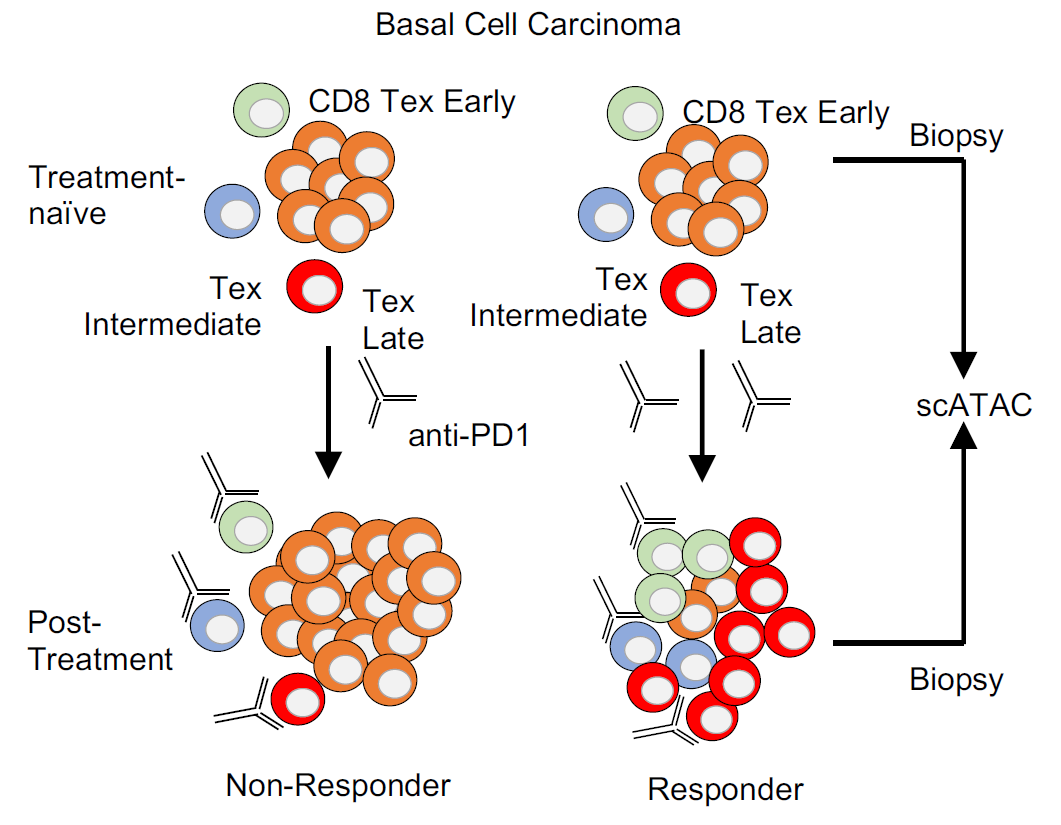
In addition, our research team has applied EpiTrace to trace immunotherapy-related T cells, kidney single cells, glioblastoma, etc., and traced the evolution of these cells from a traceability perspective.
This study is the first research-oriented long article published by Wuhan University as the first author and first corresponding unit in Nature Biotechnology. The journal Nature Biotechnology is a top tier journal in research methodology, and its published research results are usually significant innovations in pioneering research methods in the biomedical field (https://www.nature.com/nbt/collections).

Given the importance of EpiTrace, Nature Biotechnology published a Research Brief titled "Decoding cell replication age from single cell ATAC seq data" on the same day as the publication of this achievement, which gathered evaluations and recommendations from international experts and editorial teams.

This study received research funding support from the National Natural Science Foundation of China, the Basic Research Fund of Central Universities, and the Zhongnan Hospital of Wuhan University.
References:
[1] Xiao, Y., Jin, W., Ju, L. et al. Tracking single-cell evolution using clock-like chromatin accessibility loci. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02241-z
[2] Xiao Y, Jin W, Qian K, et al. Integrative Single Cell Atlas Revealed Intratumoral Heterogeneity Generation from an Adaptive Epigenetic Cell State in Human Bladder Urothelial Carcinoma. Adv Sci (2024), 2308438. doi: 10.1002/advs.202308438.
[3] Decoding cell replicational age from single-cell ATAC-seq data. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-024-02256-6
Links:
https://www.nature.com/articles/s41587-024-02241-z