


On September 14, 2022, Professor Wang Xinghuan's team of urology Department at Zhongnan Hospital of Wuhan University published the latest research results "Ultrasensitive tumor-agnostic non-invasive "in the internationally renowned academic journal Clinical and Translational Medicine detection of colorectal cancer recurrence using ctDNA methylation"[1]. This study conducted an in-depth analysis of the specific DNA methylation profile of colorectal cancer (CRC) and established a method for detecting minimal residual lesions (MRD) using postoperative circulating tumor DNA methylation features (ctDNAm). The results indicate that this method can detect colorectal cancer MRD more sensitively and specifically without the need for patient tumor tissue samples, and can be an ideal detection method for CRC risk stratification and recurrence prediction. This is another research achievement of Professor Wang Xinghuan's team in the non-invasive diagnosis and monitoring of bladder cancer based on the specific DNA methylation characteristics of urine samples[2], which uses the principle of this technology to carry out non-invasive DNA methylation detection to monitor the recurrence of solid tumors.
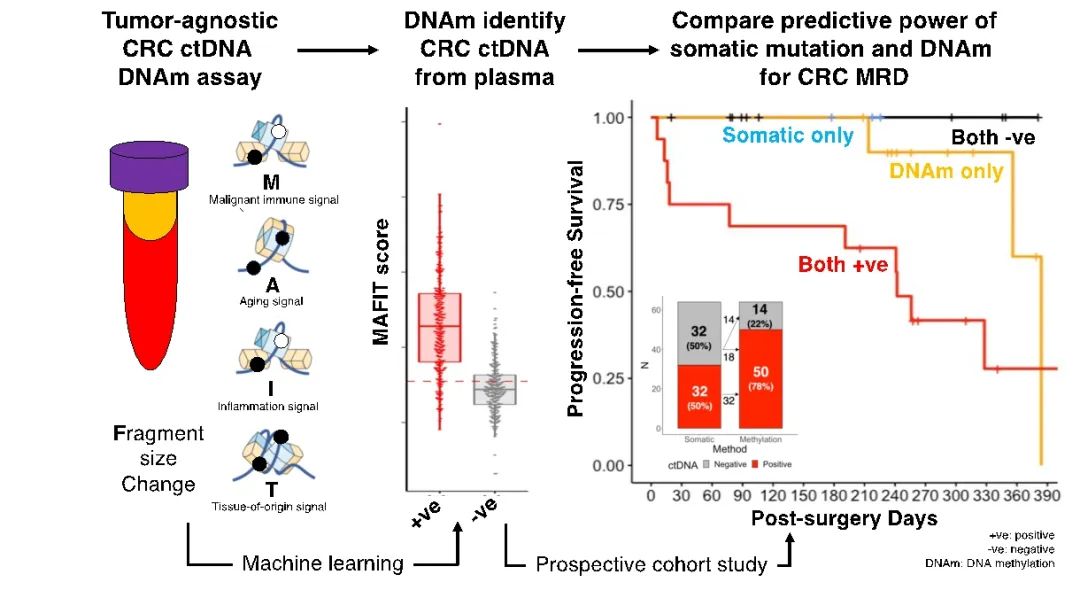
Colorectal cancer is the malignant tumor with the third incidence rate and the second cancer-related mortality in the world. The main treatment for early to mid stage colorectal cancer patients is surgical resection, and patients with high risk of recurrence require chemotherapy. At present, the difficulties faced by postoperative adjuvant therapy for early and mid stage colorectal cancer patients are twofold: firstly, how to accurately screen high-risk populations for personalized and precise adjuvant therapy; secondly, how to detect tumor recurrence earlier for early intervention. Individualized and precise treatment of colorectal cancer based on the detection of minimal residual lesions using circulating tumor DNA (ctDNA-MRD) has become a hot topic and future development trend in recent years. According to literature reports, compared to conventional clinical follow-up methods such as CT imaging monitoring, ctDNA-MRD monitoring can detect tumor residue or recurrence 8-12 months in advance[3]. For patients with negative ctDNA-MRD after surgery, routine adjuvant therapy may not be necessary to reduce unnecessary chemotherapy damage[4]. For patients who test positive for ctDNA-MRD after surgery, further treatment or replacement of treatment plans can be carried out as soon as possible.
Currently reported ctDNA-MRD tests are mainly divided into two categories: one is Tumor-informed assays and the other is Tumor-agnostic assays. The ctDNA-MRD detection based on tumor informed analysis is a paired detection analysis of tumor tissue samples and peripheral blood samples, identifying somatic mutations unique to the patient's tumor tissue, and then customizing amplification primers or targeted capture combinations for personalized ctDNA detection. Its accuracy is limited by the unique mutations of the obtained tumor tissue, and there are certain differences between different patients. The operation is cumbersome, the cycle is long, and the cost is relatively high. Meanwhile, many patients who come for medical treatment and testing are unable to provide tissue samples for surgical resection or puncture due to various reasons, which limits the applicability of this method to a wider audience. ctDNA-MRD assay based on tumor agnostic analysis is the detection of highly correlated hot spot gene mutations in a pre-screened set of tumors, without the need for primary tumor tissue, but cannot detect mutations outside the pre-selected sites. Therefore, developing more accurate and convenient ctDNA-MRD detection technology is an urgent need in clinical practice.
DNA methylation (DNAm) has different characteristics in different tumors, and the analysis of tumor-specific DNAm characteristics can be used for tumor diagnosis, histopathological origin tracing, prognosis stratification and recurrence monitoring. In the previous study, our team searched for tumor-specific DNAm signals through whole genome bisulfite sequencing, and classified tumor DNAm into five types of DNAm markers through statistical learning: DNAm signal (M) of malignancy related immune cells, DNAm signal of methylation age (A), DNA fragment distribution signal of tumor tissue (F), DNAm signal of inflammatory immune cells (I), and DNAm signal of cell origin of tumor tissue (T). In this study, the researchers used machine learning algorithms for statistical analysis of the above five types of signals, and built a MAFIT scoring model for accurate quantification of blood ctDNA for postoperative risk stratification and non-invasive recurrence monitoring of colorectal cancer patients.
In a multicenter prospective observational clinical validation cohort Challenge-CRC, a total of 280 CRC patients (64 untreated first-time patients and 216 patients after surgical treatment) were included. Untreated preoperative blood and postoperative blood were collected, and ctDNA somatic mutation analysis and ctDNAm analysis were performed on all samples. In the absence of tumor tissue, the ability of ctDNA somatic mutations and ctDNAm detection methods to detect ctDNA derived from colorectal cancer was compared, and the effectiveness of these two methods in predicting postoperative recurrence was compared. This report is the mid-term data of the queue, and the results show that the sensitivity of preoperative blood ctDNAm detection is superior to ctDNA mutations, and the specificity is not inferior to ctDNA mutations; The sensitivity and NPV of postoperative blood ctDNAm were significantly better than those of ctDNA mutation methods. Finally, researchers divided patients into low-risk, medium-risk, and high-risk groups based on their postoperative blood ctDNAm MAFIT scores. Prospective observational validation indicates that different risk groups have significantly different recurrence risks.
In summary, the performance of ctDNAm-MRD in predicting postoperative recurrence of colorectal cancer is superior to the MRD detection method based on ctDNA mutations. Through postoperative blood specific ctDNAm analysis, MRD detection can be performed more efficiently, predicting the risk of postoperative recurrence of colorectal cancer, helping clinical development of reasonable postoperative monitoring plans and follow-up intervals, and even guiding treatment interventions. In the near future, preoperative blood cfDNAm testing may also contribute to the early diagnosis of colorectal benign and malignant diseases, help reduce unnecessary invasive colonoscopy examinations, and reduce the risk of non essential surgeries.
References:
1] Xiao Y, et al., Ultrasensitive tumour-agnostic non-invasive detection of colorectal cancer recurrence using ctDNA methylation. Clin Transl Med, 2022, 12(9):e1015.
[2] Xiao Y, et al. Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNA methylation in a prospective cohort study. Clin Transl Med. 2022;12(8):e1008.
[3] Wang Y, et al., Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol. 2019, 5(8):1118-1123.
[4] Tie J, et al., Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med. 2022, 16;386(24):2261-2272.
Links: