

Recently, The team of Professor Wang Xinghuan of the Department of Urology at Zhongnan Hospital of Wuhan University published the latest research results "Non-invasive diagnosis and surveillance of Cancer" in the internationally renowned academic journal Clinical and Translational Medicine bladder cancer with driver and passenger DNA methylation in a prospective cohort study "[1]. This study systematically analyzed the specific DNA methylation profile of bladder cancer, established a detection method based on the specific DNA methylation characteristics of bladder cancer in urine samples, and carried out efficacy verification in a multi center prospective cohort. The results showed that the characteristic changes of free DNA methylation in urine could be used as an ideal biomarker for non-invasive diagnosis, prognosis and monitoring of bladder cancer.

There are two types of bladder cancer: non-invasive bladder cancer and invasive bladder cancer (NMIBC/MIBC). There are significant differences in tumor biology, treatment and prognosis. The clinical treatment of bladder cancer depends on the accurate staging of the tumor. Inaccurate staging will lead to lower survival rate or unnecessary surgery. At present, the "gold standard" for the diagnosis and monitoring of bladder cancer is histopathological examination, which must rely on invasive and costly cystoscopy biopsy or diagnostic electrotomy to obtain tissue. The sensitivity and specificity of non-invasive diagnostic methods reported so far are low, and it is impossible to distinguish between low and high grade (LG/HG) bladder cancer and NMIBC/MIBC. Therefore, developing more accurate and non-invasive diagnostic and monitoring methods that can distinguish tumor staging and grading is an urgent need in clinical practice.
Abnormal DNA methylation has been found in various tumors, and specific DNA methylation can be used for tumor detection and precise subtype classification. In previous research, the team has found that different methylation changes in bladder cancer tissue are closely related to bladder cancer pedigree and malignancy, and has located 17 bladder cancer related differential methylation regions[2]. In the training queue of this study, the team found the specific DNA methylation characteristics of bladder cancer and established a targeted sequencing analysis method for specific DNA methylation characteristics through the whole genome bisulfite sequencing of 32 bladder cancer tissues and 12 healthy control urine. Subsequently, 259 urine samples from 5 centers were tested in a double-blind manner in the validation queue. The results showed that the sensitivity of the new method based on the characteristics of free DNA methylation in urine was 100% (95% CI: 82.5% -100%) for the diagnosis of HG bladder cancer, and 62% (95% CI: 51.3% -72.7%) for LG bladder cancer. The specificity of both methods was 100%. The postoperative urine free DNA methylation characteristics are used to predict no recurrence within 180 days with an accuracy of 100%, and long-term follow-up verification is ongoing.
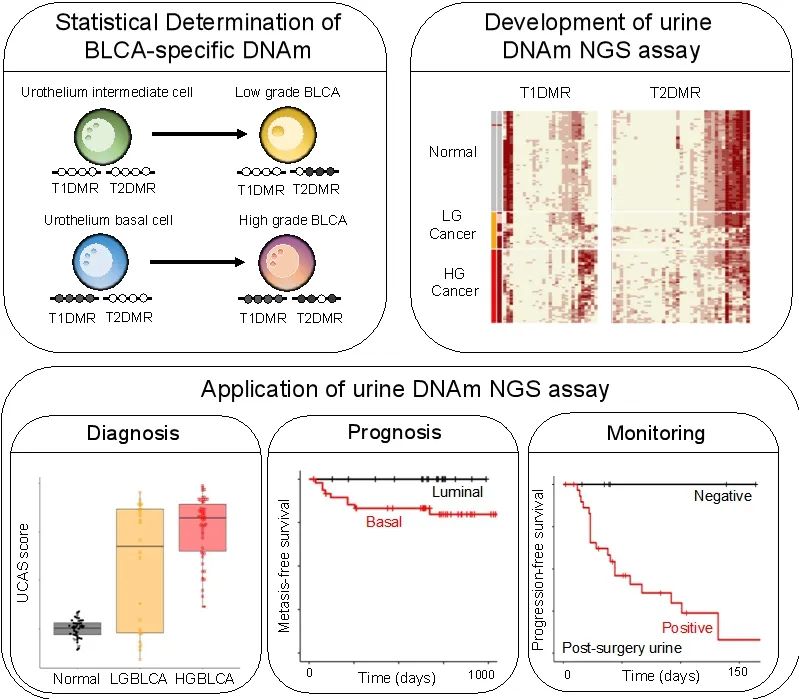
In conclusion, the detection of free DNA methylation in urine is superior to the traditional non-invasive detection method in diagnosing bladder cancer, predicting recurrence and predicting prognosis. This method helps to reduce unnecessary invasive examinations, not only alleviating the pain and economic burden of patients, but also reducing the risks associated with some unnecessary surgeries. This study not only has practical significance for the diagnosis and monitoring of bladder cancer, but also indicates the potential of similar methods to be widely used in other tumors.
References:
1. Xiao Y, Ju L, Qian K, et al. Non-invasive diagnosis and surveillance of bladder cancer with driver and passenger DNA methylation in a prospective cohort study. Clin Transl Med. 2022;12(8):e1008. doi:10.1002/ctm2.1008.
2. Xiao Y, Jin W, Qian K, et al. Emergence of an adaptive epigenetic cell state in human bladder urothelial carcinoma evolution. bioRxiv. 2021. Preprint at https://doi.org/10.1101/2021.10. 30.466556.
Links:
https://onlinelibrary.wiley.com/doi/10.1002/ctm2.1008